Direct Tests of the Energetic Basis of Abortive Cycling in Transcription
Ankit V. Vahia & Craig T. Martin, Biochemistry 50, 7015-7022, 2011.
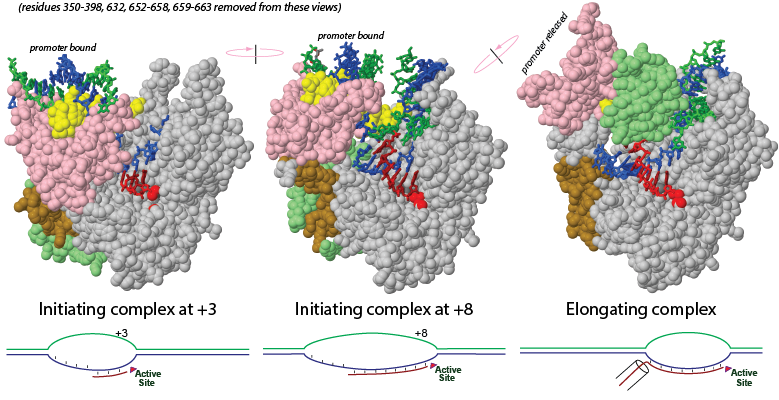
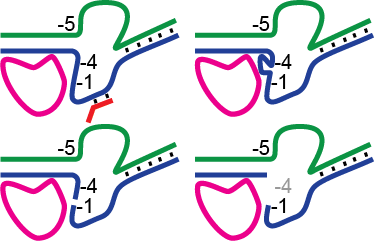
Although the synthesis of RNA from a DNA template is (and must be) a generally very stable process to enable transcription of kilobase transcripts, it has long been known that during initial transcription of the first 8–10 bases of RNA complexes are relatively unstable, leading to the release of short abortive RNA transcripts. A wealth of structural data in the past decade has led to specific mechanistic models elaborating an earlier “stressed intermediate” model for initial transcription. In this study, we test fundamental predictions of each of these models in the simple model enzyme T7 RNA polymerase. Nicking or gapping the nontranscribed template DNA immediately upstream of the growing hybrid yields no systematic reduction in abortive falloff, demonstrating clearly that compaction or “scrunching” of this DNA is not a source of functional instability. Similarly, transcription on DNA in which the nontemplate strand in the initially transcribed region is either mismatched or removed altogether leads to at most modest reductions in abortive falloff, indicating that expansion or “scrunching” of the bubble is not the primary driving force for abortive cycling. Finally, energetic stress derived from the observed steric clash of the growing hybrid against the N-terminal domain contributes at most mildly to abortive cycling, as the addition of steric bulk (additional RNA bases) at the upstream end of the hybrid does not lead to predicted positional shifts in observed abortive patterns. We conclude that while structural changes (scrunching) clearly occur in initial transcription, stress from these changes is not the primary force driving abortive cycling.
PMID: 21776950 PMCID: PMC3159029 DOI: 10.1021/bi200620q
Practical tip: nicked constructs are not useful for synthesis of RNA, since the product is an RNA-DNA hybrid - leading to single round synthesis.