Dissociation of halted T7 RNA polymerase elongation complexes proceeds via a forward translocation mechanism
Yi Zhou, Deanna M. Navaroli, Metewo Selase Enuameh, & Craig T. Martin, Proc. Natl. Acad.Sci., U.S.A. 104, 10352-10357, 2007.
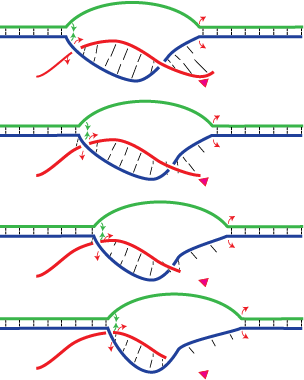
A recent model for the mechanism of intrinsic transcription termination involves dissociation of the RNA from forward translocated (hypertranslocated) states of the complex [Yarnell & Roberts (1999) Science, 284, 611-615]. The current study demonstrates that halted elongation complexes of T7 RNA polymerase in the absence of termination signals can also dissociate via a forward translocation mechanism. Shortening of the downstream DNA, or the introduction of a stretch of mismatched DNA immediately downstream of the halt site, reduces a barrier to forward translocation and correspondingly reduces the lifetime of halted complexes. Conversely, introduction of a crosslink downstream of the halt site increases the same barrier and leads to an increase in complex lifetime. Introduction of a mismatch within the bubble reduces a driving force for forward translocation and correspondingly increases the lifetime of the complex, but only for mismatches at the upstream edge of the bubble, as predicted by the model. Mismatching only the 2 most upstream of the 8 bases in the bubble provides a maximal increase in complex stability, suggesting that dissociation occurs primarily from early forward translocated states. Finally, addition in trans of an oligonucleotide complementary to the nascent RNA just beyond the hybrid complements the loss of driving force derived from placement of a mismatch within the bubble, confirming the expected additivity of effects. Thus, forward translocation is likely a general mechanism for dissociation of elongation complexes, both in the presence and absence of intrinsic termination signals.
PMID: 17553968 PMCID: PMC1965517 DOI: 10.1073/pnas.0606306104