Observed instability of T7 RNA polymerase elongation complexes can be dominated by collision-induced "bumping"
Yi Zhou and Craig T. Martin, J. Biol. Chem. 281, 24441-24448, 2006.
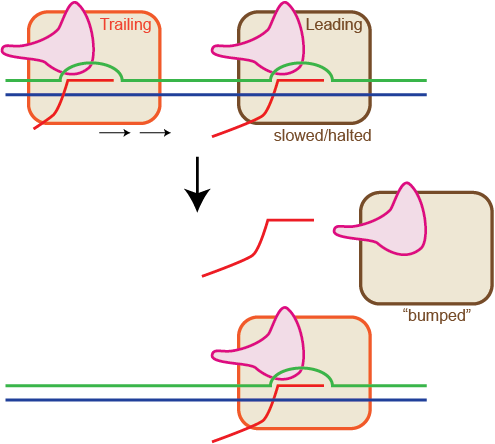
T7 RNA polymerase elongates RNA at a relatively high rate and can displace many tightly bound protein-DNA complexes. Despite these properties, measurements of the stability of stalled elongation complexes have shown lifetimes that are much shorter than those of the multi-subunit RNA polymerases. In this work, we demonstrate that the apparent instability of stalled complexes actually arises from the action of trailing RNA polymerases (traveling in the same direction) displacing the stalled complex. Moreover, the instability caused by collision between two polymerases is position dependent. A second polymerase is blocked from promoter binding when a leading complex is stalled 12 bp or less from the promoter. The trailing complex can bind and make abortive transcripts when the leading complex is between 12 to 20 bp from the promoter, but it cannot displace the first complex since it is in a unstable initiation conformation. Only when the leading complex is stalled more than 20 bp away from the promoter, can a second polymerase bind, initiate, and displace the leading complex.
PMID: 16816387 DOI: 10.1074/jbc.M604369200
Practical tip: always use an excess of promoter DNA in your reactions.