RNA - Opportunities & Challenges
At this point, mRNA vaccines need no introduction; the dramatic success of the SARS-CoV-2 mRNA vaccines has been nothing short of phenomenal. More vaccines (including personalized immunotherapies for cancer), and critically, an even broader class of mRNA therapeutics lie right behind. From mRNA protein replacement therapies for diseases like cystic fibrosis and methylmalonic aciduria, to applications in surgery, delivery of mRNA as a therapeutic is ready to revolutionize medicine.
The manufacturing of mRNA on >100 kg scales has also been nothing short of phenomenal. To enable emerging biomedicines, such manufacturing needs to be reduced in cost and distributed world-wide, democratizing access. All of the mRNA vaccines to date have been synthesized using the enzyme T7 RNA polymerase, coincidentally the enzyme that the Martin lab has studied for decades.
Challenges
One of the primary barriers in the synthesis/manufacturing of RNA is the unintended production of double stranded RNA (dsRNA) impurities. While these impurities are not harmful in themselves, our bodies have evolved a first line defense system (the innate immune response) that sees double stranded RNA as evidence of a viral attack (many viruses that infect us are double stranded RNA viruses). Tricking the immune system into thinking it is being attacked can be good for vaccines (though too much is not good!). For other purposes, this response, which triggers inflammation, can destroy the utility of an otherwise game-changing therapy.
Double stranded RNA is not an intended product of RNA synthesis and batch-synthesized RNA must be purified to reduce dsRNA to acceptable levels. We have a different idea: change the reaction so that dsRNA is not produced in the first place! Read more here...
dsRNA is an unintended byproduct of current manufacturing. Long stretches of dsRNA are not encoded in therapeutic RNAs; they are generated by the enzyme that synthesizes the RNA in vitro under conditions to generate high yields of product.
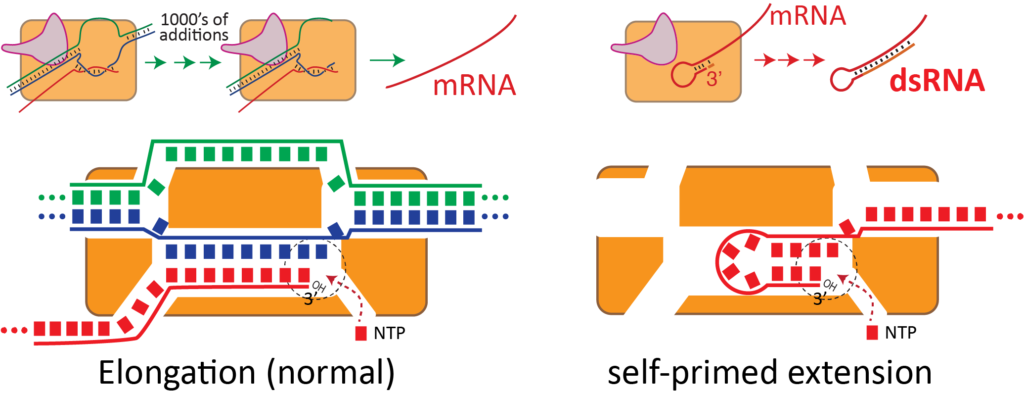
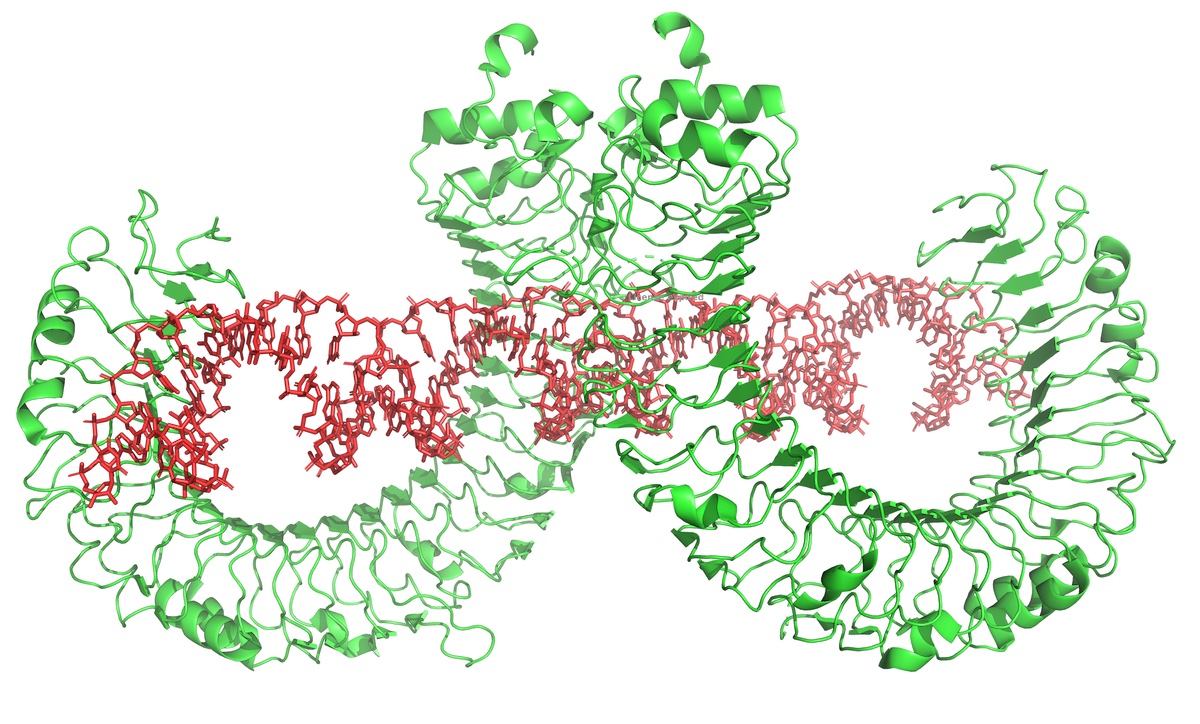
As shown above, the same active site that efficiently synthesizes RNA from a dsDNA template can also re-bind the product RNA, to now use the RNA itself as a template for extension. This is a complex, distributed process that generates a wide variety of dsRNA lengths at the end of the RNA. These regions then bind to Toll-like receptors.
Although immune stimulation increases with dsRNA length, as shown above, 40 bases is sufficient to trigger an undesired response. Purification can reduce the longer lengths, but separating a 4,000 base desired RNA from a 4,040 base RNA+dsRNA extension impurity is a herculean task, as illustrated at scale below.

We aim to eliminate the rebinding that generates dsRNA. We have some published approaches for helping, but our ultimate goal is to remove the (initial/desired) RNA from the reactor as soon as it is made. In this way, it simply cannot rebind to the enzyme. The key is functional co-immobilization of both DNA and enzyme to allow a continuous flow reactor.
Opportunities in RNA
The following presents only a sampling of applications for RNA in human health. And it should probably be updated weekly! The field is advancing rapidly, and we aim to enable that advancement!
Vaccines. The phenomenal success of the SARS-CoV-2 mRNA vaccines demonstrates the power of mRNA as a treatment modality. 1,000's of kilograms of mRNA were produced on a very short development and production timeline. New vaccines for other diseases are in clinical trials.
Vaccines can tolerate low levels of dsRNA impurities, which can serve as an adjuvant, but since the responses described above can lead cells to down-regulate translation, too much can lead to dosage issues.
Gene replacement. mRNA vaccines need no introduction, but there is a wealth of opportunity in the much broader RNA space. mRNA therapeutics aims to replace protein biologics and has a number of significant advantages. Most predict an explosion in therapies that deliver mRNA to cells for expression.
These approaches need synthetic mRNA in lengths from 1,000 to 12,000 bases. T7 RNA polymerase is the only practical system for creating these RNAs. Purity requirements vary, but the goal should always be maximal purity.
CRISPR introduction. An excellent overview of the wide range of CRISPR approaches can be found at AddGene. While nature uses a two-piece guide RNA (gRNA) system, most current applications use a single, combined RNA: sgRNA (read more here). Starting lengths for sgRNA are about 100 bases, though some novel approaches are using expanded sgRNAs. Although chemical synthesis of 100 base sgRNAs is possible, yields and purity decrease with increasing length (a chemical coupling efficiency of 98% predicts a 13% yield; the remaining 87% must be removed through purification), which further reduces yield). In vitro transcription is an attractive/preferred alternative.
CRISPR technologies - non-editing. Many are aware of the abilities for CRISPR Cas9 gene editing, to permanently treat genetic disorders. While progress in that field continues (see below), there are a wide variety of approaches emerging that use inactivated Cas9 (dCas9), exploiting its precision targeting abilities. This allows for therapeutics that can target modulation of gene expression to treat a wide variety of diseases, without inducing (permanent) changes in cellular DNA.
Although chemical synthesis can generate RNAs up to about 100 bases, yields are very low and extensive purification is required.
CRISPR technologies - editing. The initial promise of CRISPR technologies is gene editing. From initial approaches using active Cas9, advances have focused on the precision of targeting (a key attribute of the CRISPR system) and the precision of editing. Recent advances in the latter have, as above, used dCas9 to deliver other editing approaches (see recent cleavage-free dCas9-based editor).
Many of these approaches involve gRNA's of increasing lengths.