Elongation complex stability
Stability of an RNA polymerase during elongation of RNAs 10,000 bases in length or more is critical, since unlike distributive DNA polymerases, once RNA is released from an elongation complex, it can rebind and resume extension where it left off on the DNA. All RNA polymerases have about an 8 base pair RNA DNA hybrid in their elongation complexes; as a base is added at the 3′ end, an upstream base dissociates (resolves) from the hybrid and enters an RNA exit channel, before exiting the protein.
Why 8 base pairs?
RNA polymerases, from the small single subunit enzyme here, to much larger and unrelated RNA polymerases from bacteria and humans, all maintain an about 8 base pair RNA-DNA hybrid during elongation. Indeed, initial transcription is designed to stabilize the system until this length is achieved. Why 8 base pairs? An 8 base duplex is not stable in solution (though it may be when associated with the polymerase) and it has been hypothesized that 8 base pairs is the length required to establish a topological locking of the RNA around the template strand DNA.
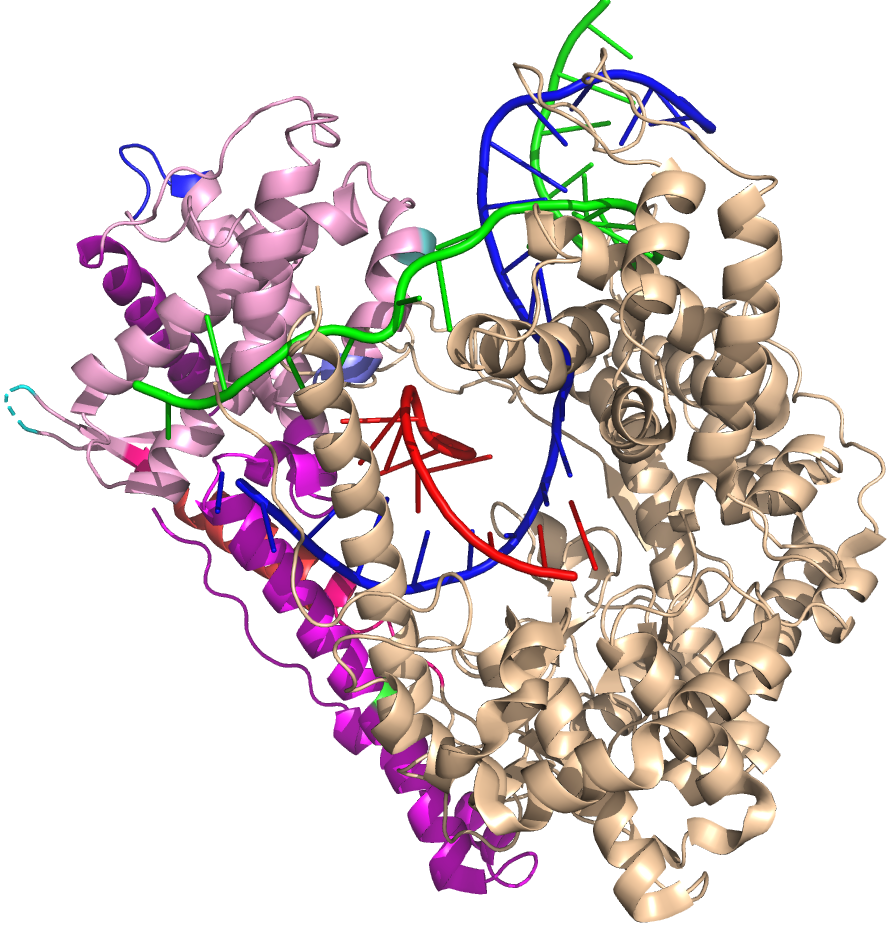
Note in the above that the red RNA strand, with its 3' end in the active site (rightmost base) and forms an 8 bp RNA-DNA hybrid. The remainder of the 5' end of the RNA exits the polymerase towards the "back" of this structure as shown. As such, the green nontemplate strand of the DNA and blue template strand cannot come together because their reannealing is blocked by the RNA. The transcription bubble is locked open. This is independent (and in addition to) any thermodynamic stability of the RNA-DNA hybrid (which is expected to be only somewhat more stable than the reannealed DNA-DNA duplex).
Nucleotide Addition Cycle - the Brownian Ratchet
Elongation in DNA polymerases, RNA-dependent RNA polymerases, and RNA polymerases all follow a common theme. The chemistry of nucleotide addition is a phosphoryl transfer reaction, where the 3' OH of the last added base attacks the (alpha-phosphate of) the 5'-triphosphate on the incoming substrate base, releasing pyrophosphate (di-phosphate).
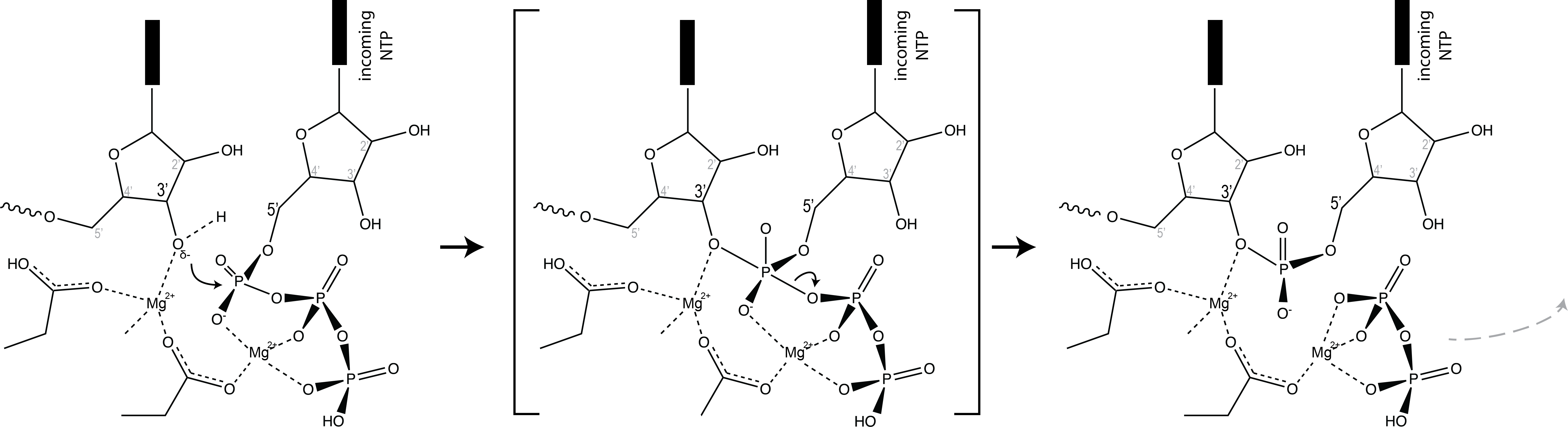
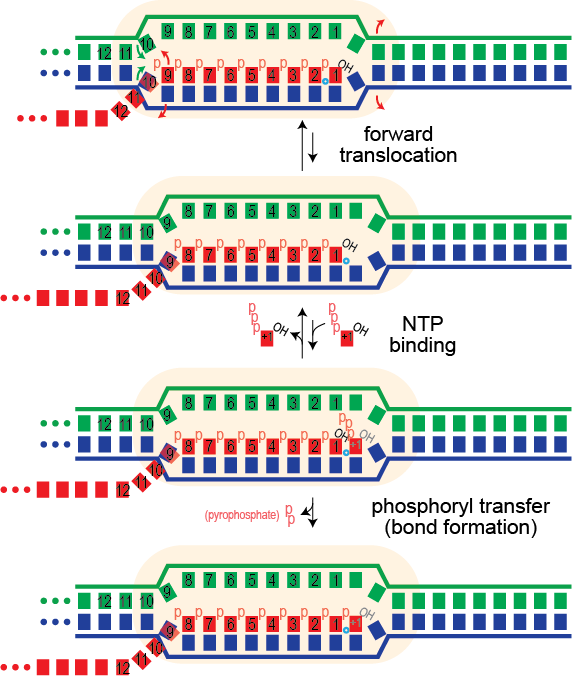
Elongation proceeds via a Brownian ratchet mechanism (nicely described by Sousa et al for T7 RNA polymerase). For this family of polymerases, the system can exist in two states: pre-translocated and post-translocated (and the enzyme can exist in two states: a closed state and an open state). The pre-translocated state exists just after addition (bond formation) of the previous base. In this state, the just-added base sits in the position where the next incoming base needs to bind. The complex then sets up an equilibrium between that state and the post-translocated state. To get from pre- to post-translocated, the enzyme moves forward by one base (or equivalently, the nucleic acid framework moves back by one base).
This family of polymerases is characterized as having a "fingers-palm-thumb" structure. The fingers domain is dynamic and can exist in "open" and "closed" states (a domain motion). The story at right is a bit more complicated, as the transition between these two states blocks and then allows phosphoryl transfer.
In the first panel below (1S77), an RNA base (red) has just been added, complementary to the DNA (blue) - enclosed by dotted lines. A tyrosine (Y639) stacks on that base pair to stabilize it. The enzyme can then translocate forward (or equivalently, the RNA-DNA moves backward) by one base. At the same time, the fingers domain moves from the "closed" to the "open" state. Y639 still stack on the last base pair, but has moved with them (1H38). For an excellent movie illustrating this, see Yin & Steitz 2004 - scroll to the botton.
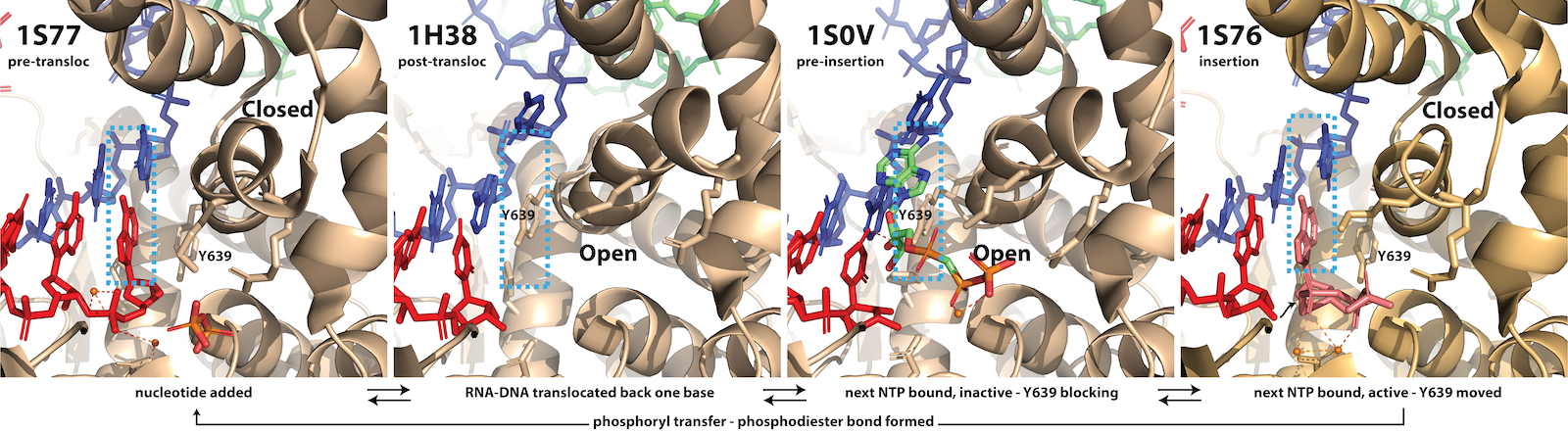
An NTP can then bind (primarily through interactions of its triphosphate with the enzyme), but cannot pair correctly, as Y639 is blocking it (1S0V). In the last panel below, a correctly base-paired NTP can effectively compete with Y639, pushing it aside, driving the fingers domain into the "closed" state (1S76). Note that all of these intermediates are in dynamic equilibrium, going back and forth via thermal (Brownian) motion. Finally, (only) in the correctly paired, closed intermediate, the 3' hydroxyl of the last added base is poised to attack the alpha-phosphate of the incoming NTP, driving phosphoryl transfer (formation of the phosphodiester bond). The process is now back at (1S77) and cycle repeats. Since phosphoryl transfer (covalent addition of the added base) is generally irreversible, it serves as the "pawl" in the Brownian ratchet, preventing excursions backward.
Thermal energy can drive off-pathway translocation(s)
Note that, starting from the post-translocated state, it is possible for the complex to (forward) translocate another time: hyper-forward translocation. This moves the 3'-OH of the last added base still further back, out of the active site. As such, a hyper-forward translocated species is catalytically inactive and must backward translocate to return to the above functional path.
The cartoon below shows a single hyper forward translocation step (the bases are numbered arbitrarily to allow one to follow the movement). Note that the polymerase and the melted bubble move forward by one base. Movement of the bubble involves:
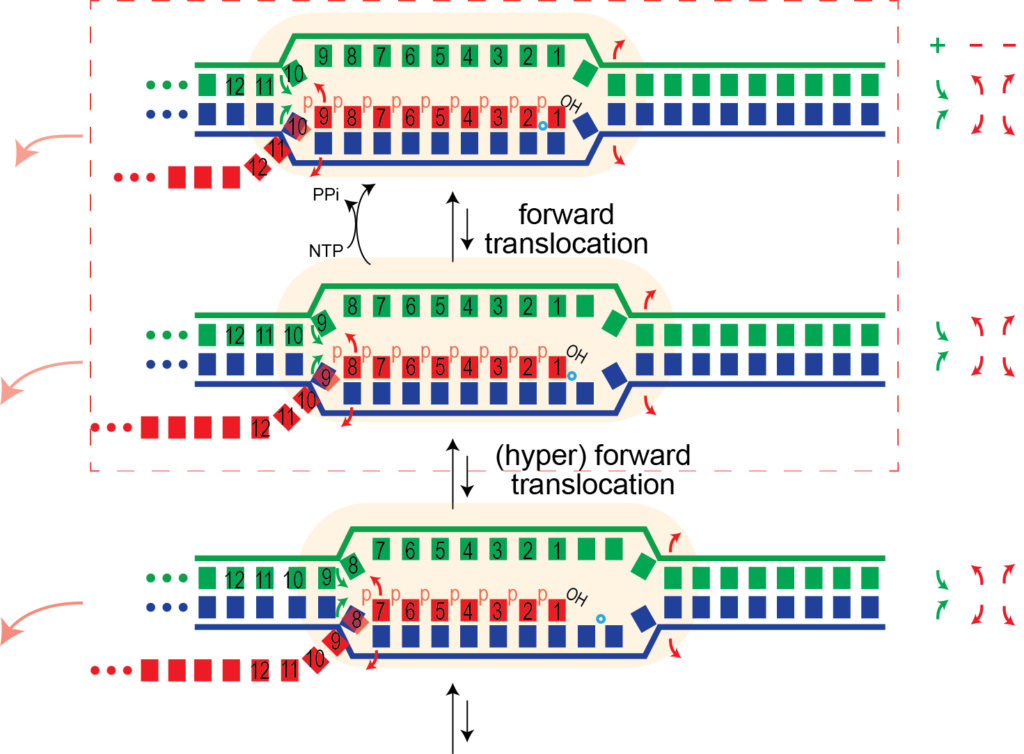
- melting of one base pair in the DNA (right-most red arrows)
- melting of one base pair in the RNA-DNA hybrid (leftmost red arrows)
- reannealing of one base pair upstream
The first two above are unfavorable, balanced only partly by the third favorable process. A "pushing" or "bumping" force from the left (upstream) could be enough to drive this process - see below.
This one base pair movement is probably not enough to unthread the topological lock (otherwise undesired RNA release might happen during normal elongation). But at some point, perhaps following 4-5 hyper forward translocation events, the RNA may be sufficiently unthreaded to now be able to dissociate, allowing the bubble to collapse, releasing the polymerase and the RNA.
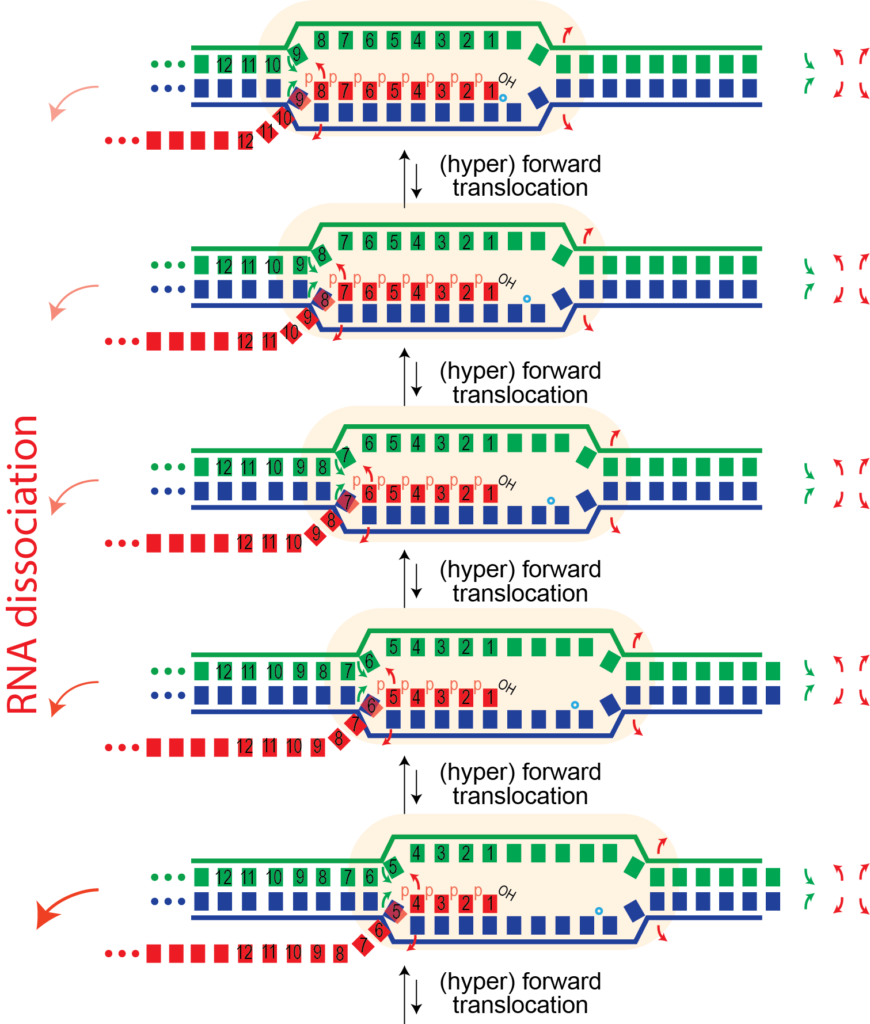
Why doesn't this happen more often? Perhaps it does, but not with enough forward translocation steps to reach the point of critical instability. Note that each hyper forward translocation event is reversible - the polymerase can reverse forward translocate.
Runoff transcription
While an elongation complex is very stable, even when halted by lack of substrate, when the polymerase reaches the end of a template, the complex becomes very unstable and the RNA dissociates.
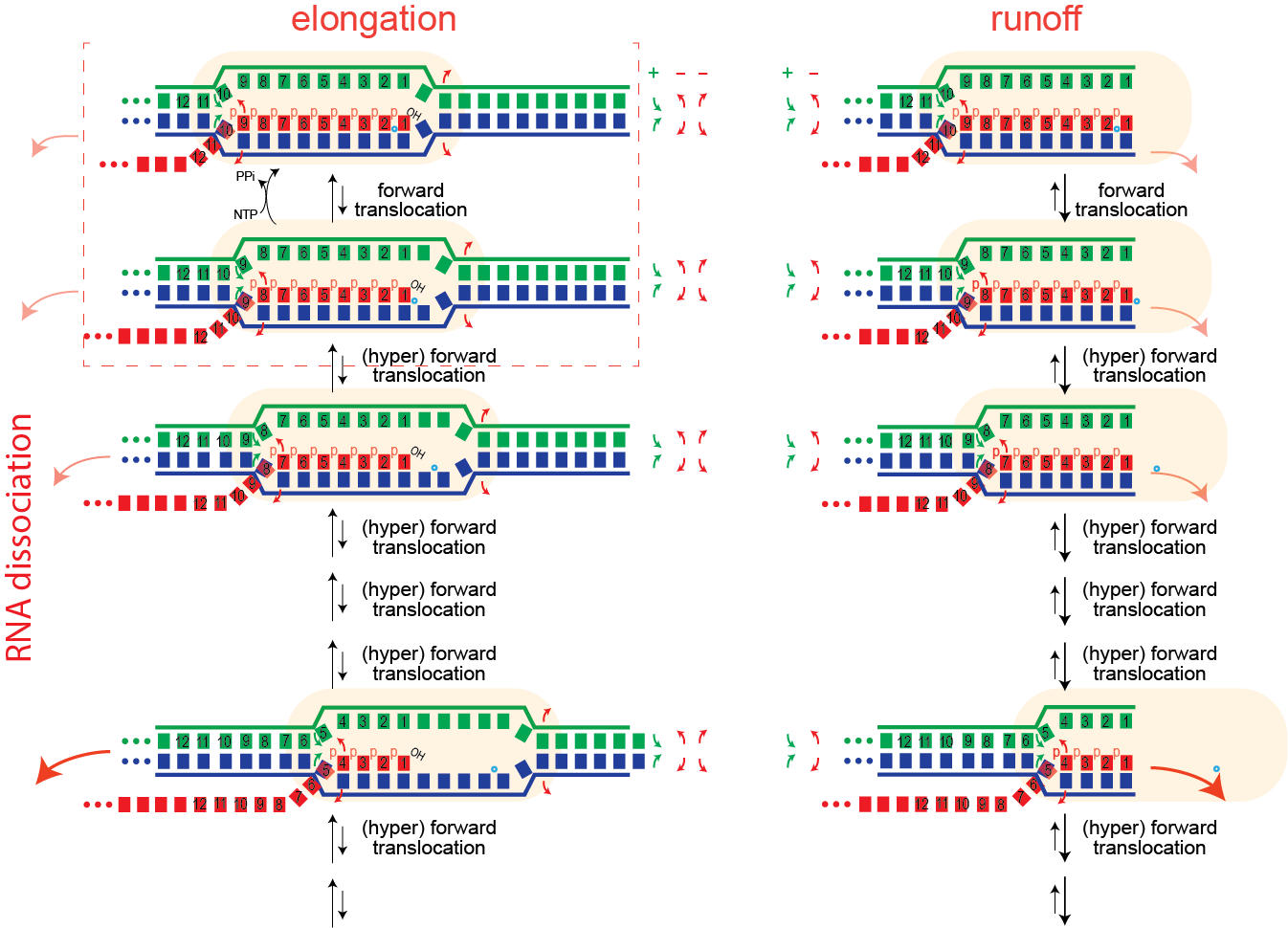
Termination (Type I - hairpin-dependent)
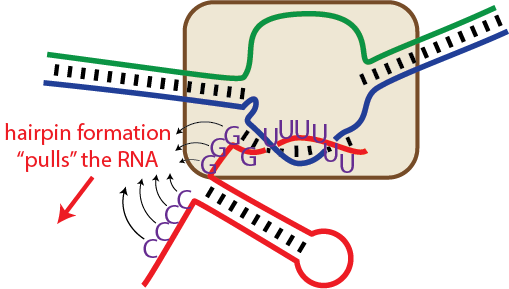
It is generally accepted that in Type I termination, formation of a hairpin in the RNA exiting the polymerase drives hyper forward translocation to unthread the lock, as above. This can be thought of (equivalently) as pushing on the polymerase or pulling on the RNA).
Termination requires a hairpin some number of bases upstream of the termination site, but it also requires a run of encoded Us in the RNA.
Note that termination is determined by a kinetic competition between elongating forward and RNA release. Thus, more important than the overall thermodynamic stability of the hairpin is the rate of formation of the hairpin and of the key bases at the closing end of the hairpin.
The run of U's may also allow slippage of the RNA relative to its DNA template, providing an alternate route to shortening of the hybrid and unthreading of the lock.
For a very nice review of termination in bacterial RNA polymerase see Peters, Vangeloff & Landick (2011). Although the polymerases are very different, the fundamental mechanisms of termination are very similar.
Termination (Type II - no hairpin)
T7 RNA polymerase also recognizes another sequence (ATCTGTTTTxTTxx)), which leads to pausing and/or termination. This sequence was originally discovered in the human preproparathyroid hormone gene and so is also known as the PTH terminator. A related sequence (TATCTGTTttctt) plays a functional role at the concatemer junction in the T7 phage genome and serves as a pause/termination site. See MacDonald et al 1994, Hartvig and Christiansen 1996, Ma et al 2005, and Nayak et al (2008).
("Termination" by) bumping by a trailing RNA polymerase
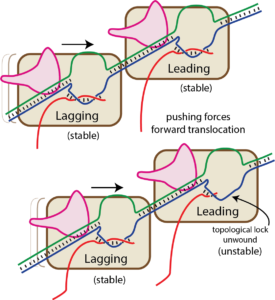
Bumping by a trailing (lagging) RNA polymerase can displace a leading RNA polymerase (this is true for the T7 family, but not for the multi-subunit family of RNA polymerases). We have proposed that this happens by the trailing RNA polymerase “pushing” against the leading RNA polymerase, which can lead to (hyper) forward translocation (that is, without incorporating a base into the RNA). Hyper forward translocated complexes have removed the 3′ base (and the associated 3′ hydroxyl) from the active site and so are inactivated. Further hyper forward translocation reduces the size of the RNA-DNA hybrid and unthreads the topological lock. The resultant instability allows the complex to dissociate, ending transcription irreversibly.
How does an elongation complex come apart in the middle of the DNA, as occurs during a process called termination?
For this we need to "unthread" the topological locking of the RNA around the DNA. We believe this happens through a process called (hyper) forward translocation.
As you can see in the "bumping" cartoon above, a trailing RNA polymerase can "push" a stopped RNA polymerase, sliding the stopped polymerase forward. But in order for that to happen, the melted bubble must move in accordance (the structure of the complex requires this).
T7 RNA polymerase is a motor protein
The above characterizations are consistent with T7 RNA polymerase as a powerful motor protein, Force studies have supported the kinetic models shown above. In particular, force studies have shown that at moderate concentrations of substrate NTPs, the velocity of an elongating polymerase is independent of NTP concentration.
T7 RNA polymerase can function as an RdRp
More to come here.
Mutant protein claims. Various phylogenetically-related polymerases and mutants of T7 RNA polymerase have been reported to reduce the RdRp activity of the enzyme.
S43Y (2021). This single mutation has been shown to eliminate termination at a class I terminator and reduce dsRNA formation (RdRp activity). This residue is located at a "hinge" that connects helices C1 (28-41) and C2 (46-55). In the initiation configuration, these two helices lie "bent" relative to each other. In the transition to elongation, the helices become almost co-linear.
G47A + 884G (2022). Each mutation individually showed a modest impact, but together had a larger effect. A variety of C-terminal additions (884G represents a Gly added to the native C-terminus of the enzyme (883) show some reduction. Note that the C-terminus alternates between two configurations: exposed to solvent and buried in the active site. The single mutants S43E, S43E, S43R, G47A, and G47L also showed some reduction. Curiously, RNa produced from A260R showed reduced immunogenicity, but a minimal effect on 3' heterogeneity.
Codexis (2024). A broad set of claims across a broad set of mutants makes analysis difficult: see patents here, here, here, and here. Caveat emptor.