Promoter Binding
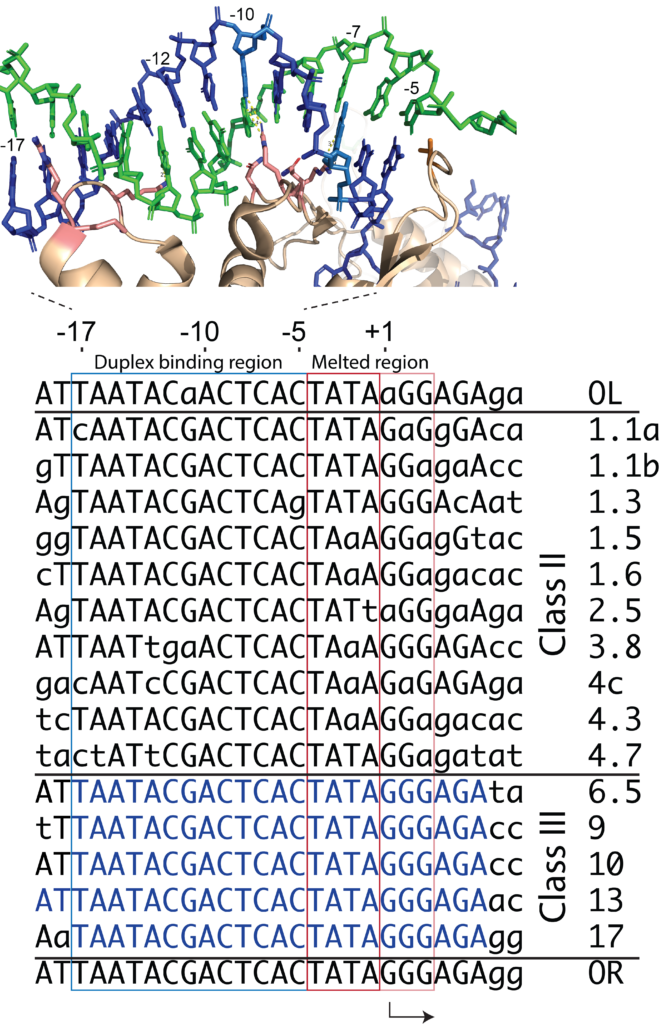
The viral genome of the bacteriophage T7 contains 17 recognized promoters. They are divided functionally between the class II and class III promoters. The Class III promoters are generally stronger and represent the consensus promoter sequence. Most researchers use this consensus sequence.
Consensus promoter: TAATACGACTCACTATAGG
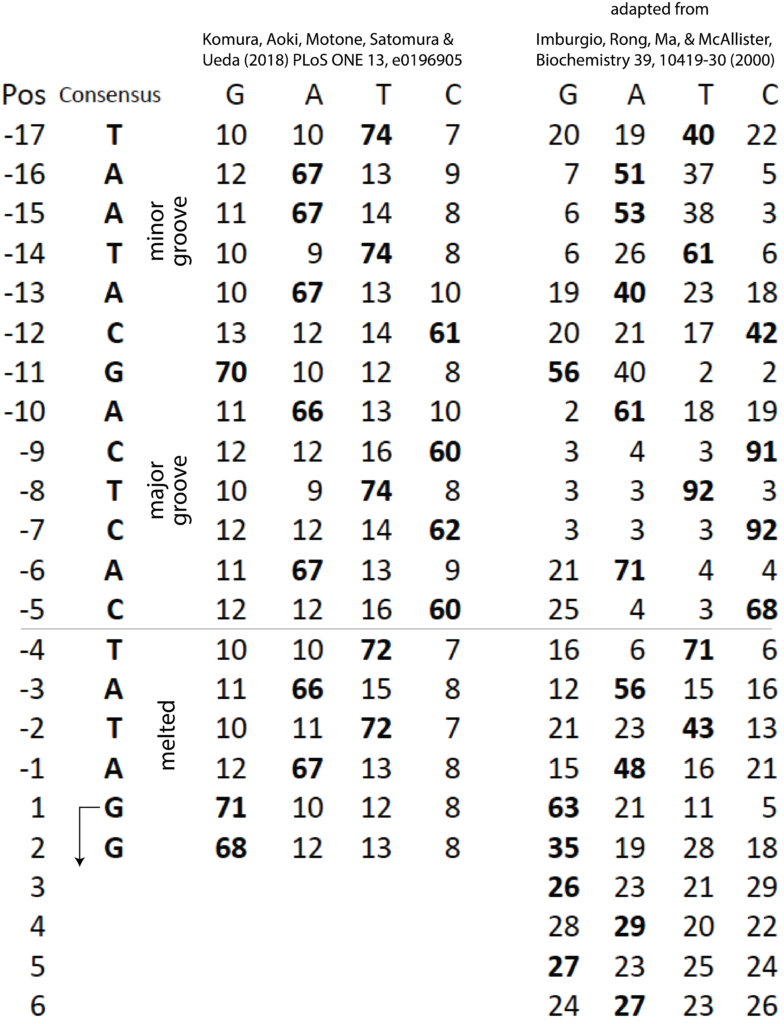
Positions -10 to -6 are strictly conserved in all 17 promoters in the 40 kilo base T7 phage genome, as shown at right. (actually, with one exception, the sequence is conserved from positions -10 to -3).
It is important to understand that the promoters are divided loosely into two regions. DNA bases between position -17 and -5 remain duplex throughout transcription (as shown at right) and drive initial interactions with the polymerase. That binding energy is used to melt the region of the DNA from position -5 to about +3 (transcription starts at position +1; there is no "0" position).
Within the duplex binding region, some bases are more critical than others. As shown in the crystal structure, the enzyme interacts with a central major groove from about positions -12 to -7. These contacts are the strongest and provide the most sequence specificity among the T7 family of RNA polymerases.
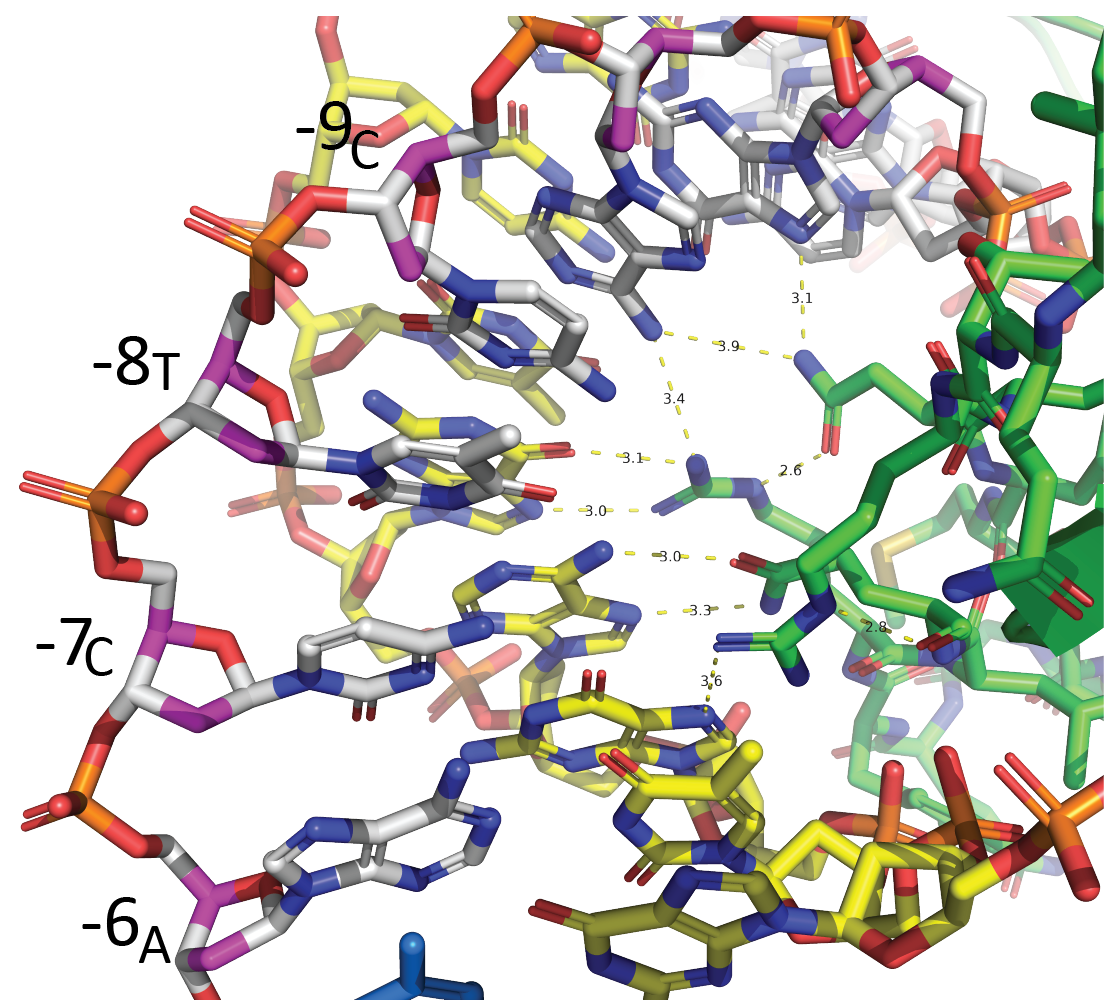
Much of this interaction derives from interactions with the specificity loop, a region of the C-terminal domain that rests on the N-terminal domain. In the figure shown at left, you can see that specificity loop side chains make sequence-specific contacts with major groove elements from positions -6 through -11 (the left-most strand, in white, is the nontemplate strand).
Upstream of the central major groove interactions, the enzyme interacts with the duplex minor groove from about positions -17 to -13. Here the protein "reads out" negative charge in the minor groove, using Lys and Arg residues from the N-terminal domain.
The initially transcribed region lies from position +1, the state site for transcription, through about position +8, where the complex transitions from a promoter-bound initially transcribing complex to a promoter released elongation complex. Promoter release involves loss of the upstream duplex interactions, a large structural change in the enzyme (in which an RNA exit channel is formed), and the displacement of the 5' end of the RNA from the RNA-DNA hybrid into the RNA exit channel.