Intro to Nucleic Acids
Under development
A key concept in nucleic acids as they exist in biology is the concept that the Watson-Crick base pairs are isosteric. In terms of satisfying H-bonding optimization, there are a number of good ways of combining two bases. But we see only AT, TA, and GC and CG pairs in DNA. That's because double stranded DNA is synthesized by DNA polymerase. That is, one enzyme with a single active site must be able to correctly select for AT or TA or GC or CG, but not other combinations (GU, for example). How does DNA polymerase select for this? Answer: not by "reading out" anything particular about any single base. Rather, DNA polymerase has a single active site that catalyzes phosphoryl transfer (base addition). Watson-Crick base pairing positions the incoming base into that active site.
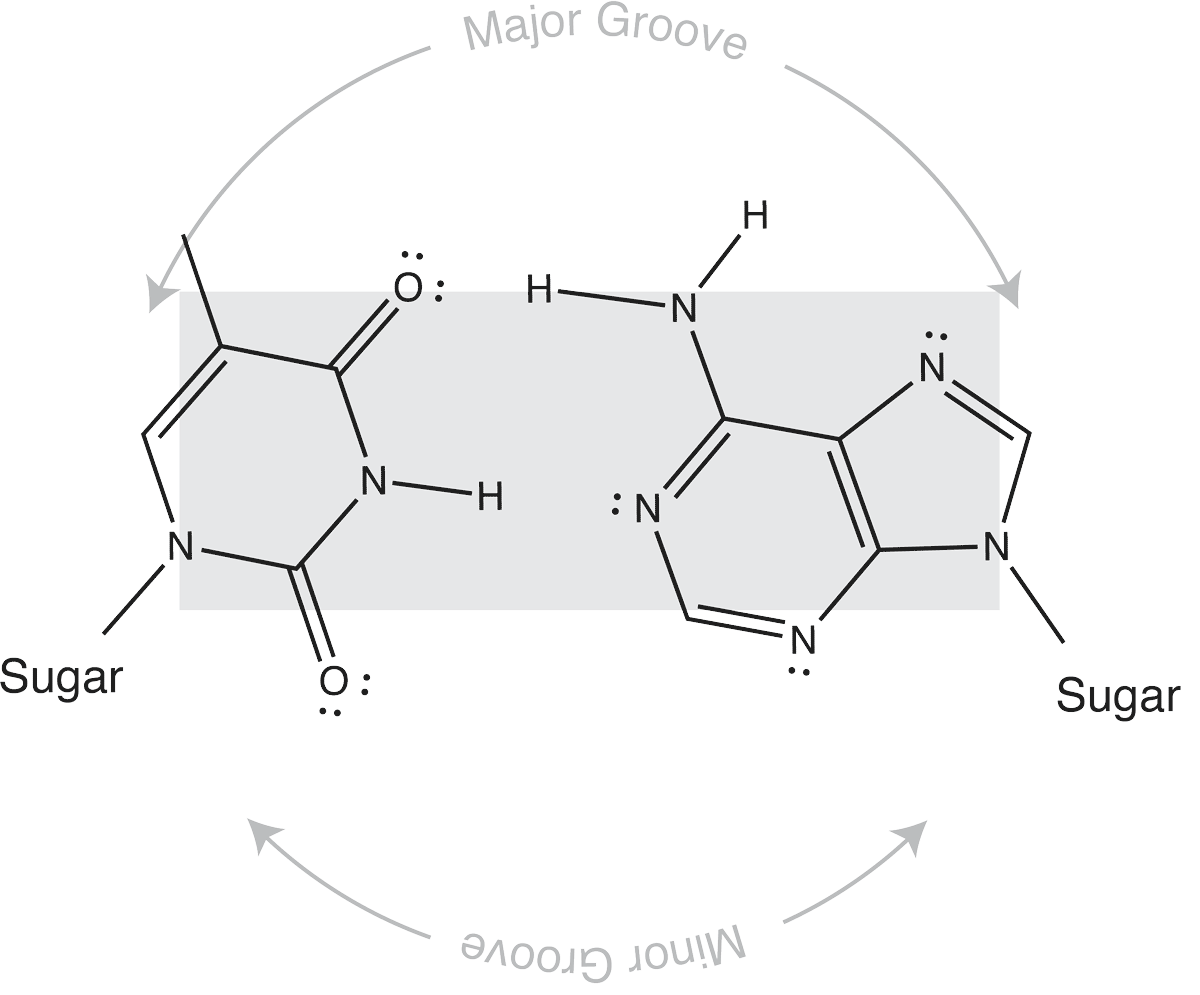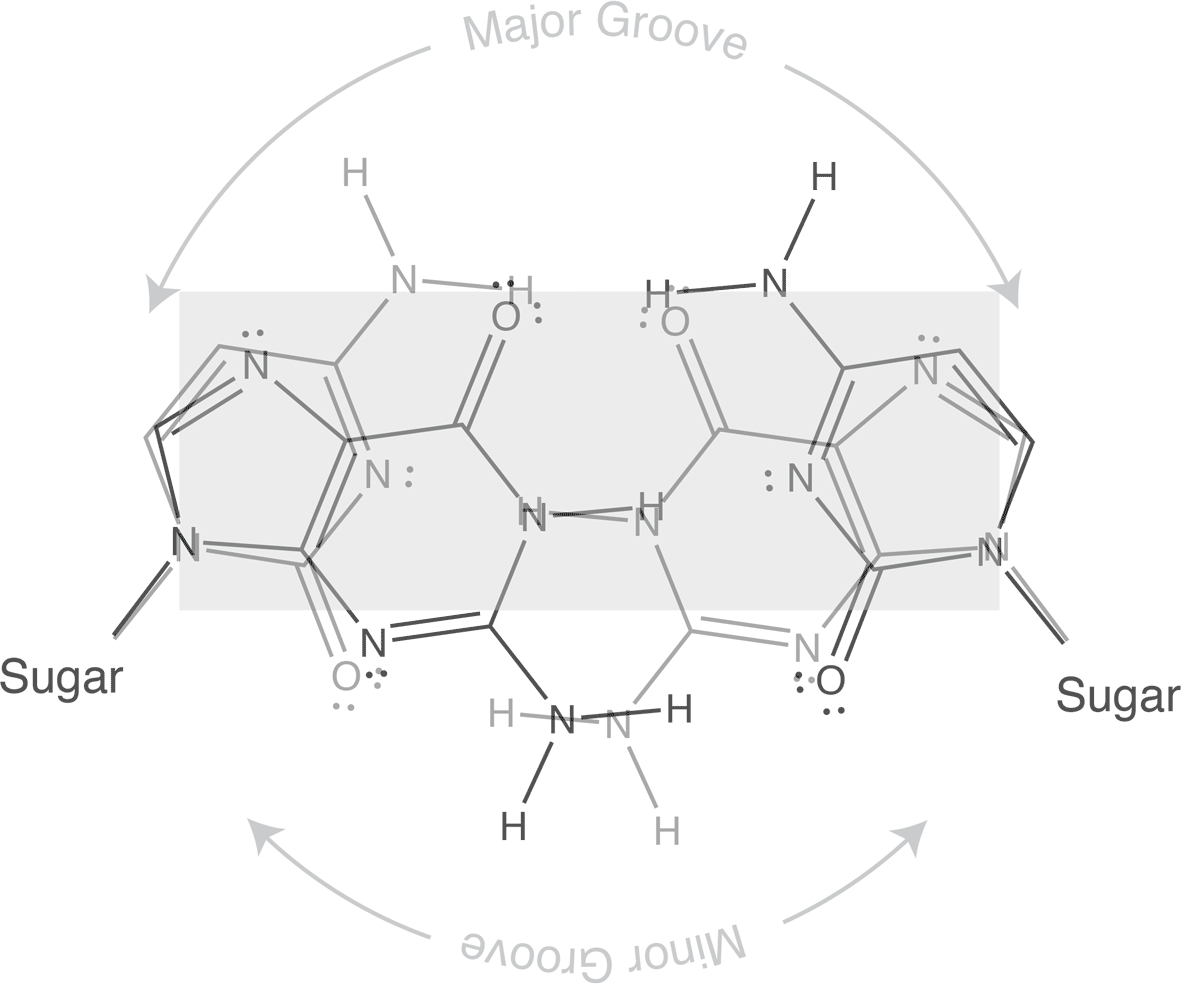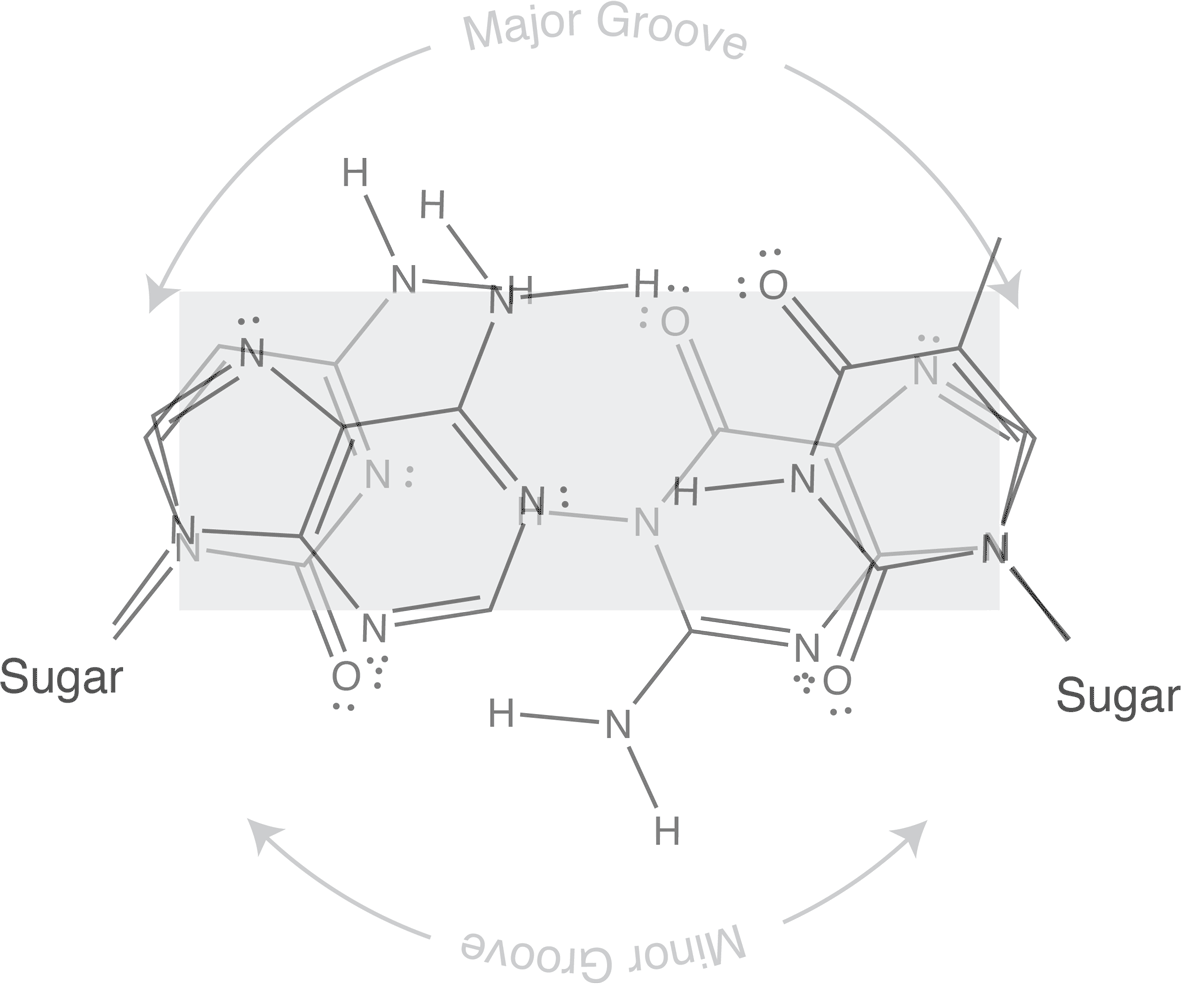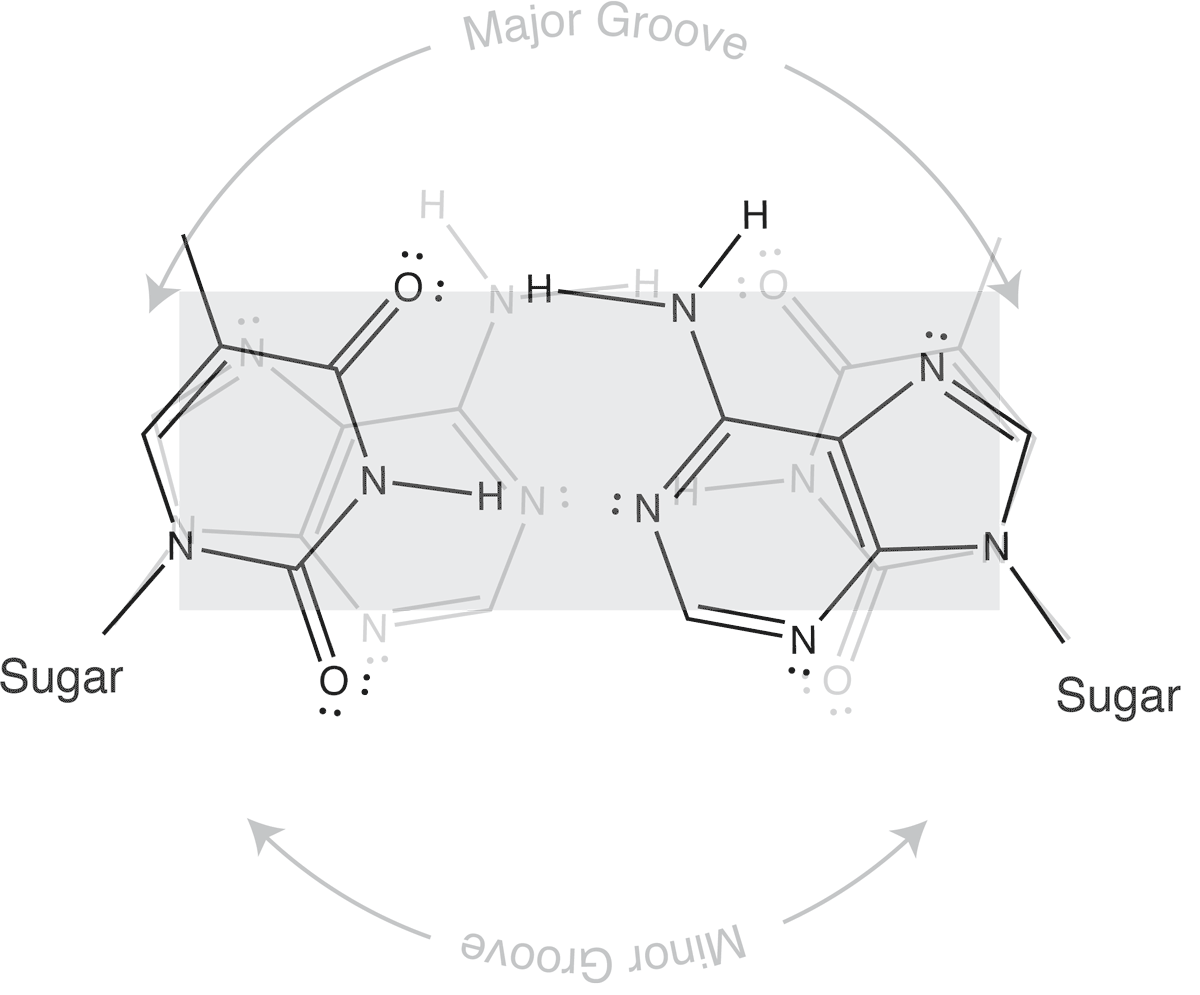
Which brings us to the concept of isosteric base pairs. This animation shows that the four Watson-Crick base pairs fit nicely into exactly the same space (they are isometric). That means that each of AT, TA, GC, and CG directs the sugar into exactly the same position in the polymerase active site, poised properly for catalysis. So if the base pair on the left is the "templating" base, the base pair on the right needs to be positioned with its sugar 5' triphosphate positioned to stabilize the nucleophilic attack by the 3' hydroxyl of the last incorporated base.
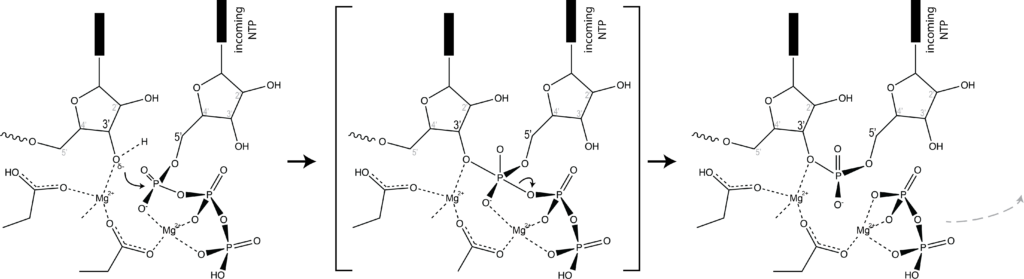
Other perfectly good base pairs move that sugar (and its 5' triphosphate) into the wrong place - catalysis does not occur!
The isosteric nature of Watson-Crick bases explains why a DNA duplex is so uniform in the path of its sugar-phosphate backbone. That path is the same regardless of the sequence. It is also the basis for recognition in exonucleolytic proof-reading.
Are all base pairs in biology Watson-Crick? No! RNA is synthesized in exactly the way described above, but structured RNAs then fold up into base paired structures using energetics alone, not an enzyme, to dictate the folding. A U added by polymerase (and encoded by an A) can pair up with a G added in another place in the sequence (encoded by a C), to form a GU base pair.
The DNA/RNA duplex
DNA and RNA adopt different duplex forms, but the principles above are retained. In standard B-form or A-form duplexes, the bases still come together Watson-to-Crick, and with their sugars on the same side of the base pair.
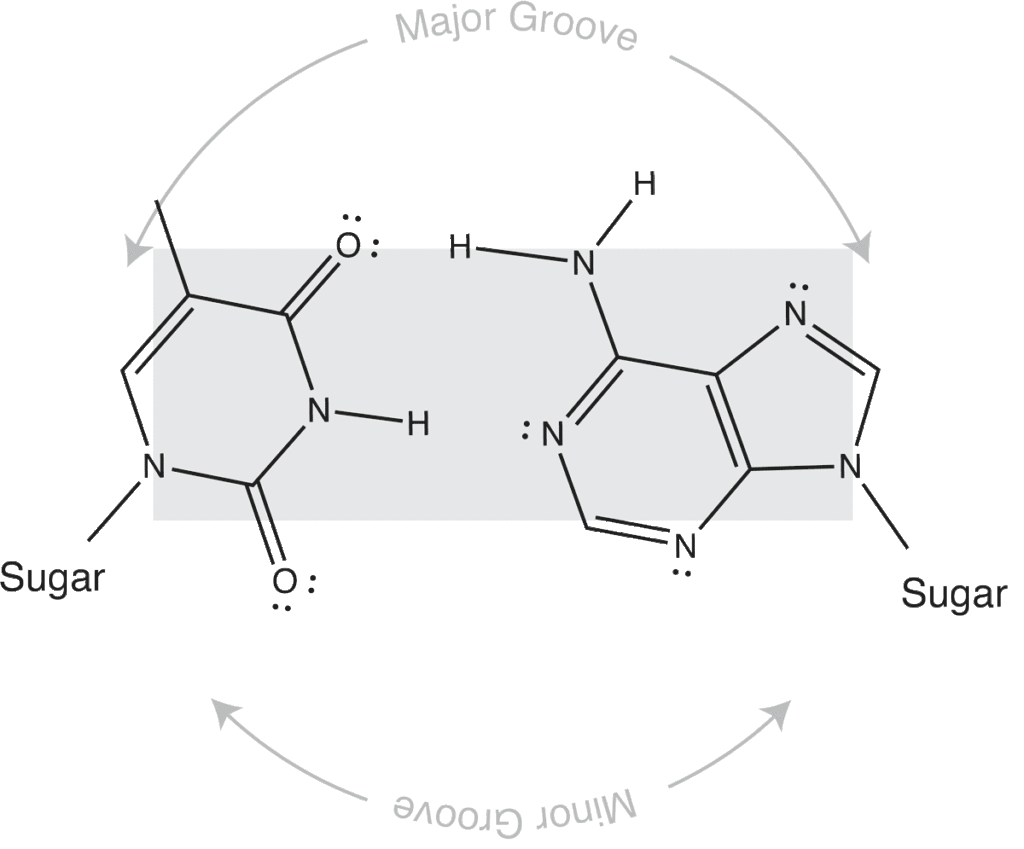
Major and minor grooves are defined by the sugar positions.
Note that the "circular path" from sugar connection to sugar connection can follow a long path or a short path. The long path defines the major groove. The short path defines the minor groove. In (B-form) DNA, both are relatively shallow and the major groove is wider than the minor groove. In an (A-form) RNA duplex, the major groove is narrow and deep, while the minor groove is very shallow and wide.